
| Project |
| Pep2Path |
| Download |
| Documentation |
| Installation |
| Tutorial |
| FAQ |
| Contact |
Pep2Path: Automated mass spectrometry-guided genome mining of peptidic natural products
Pep2Path is a simple and efficient algorithm that automates the identification of biosynthetic gene clusters for peptides analyzed by tandem MS approaches, by matching mass shift sequences from mass spectra to the gene clusters that most likely encode the corresponding peptide. Thus, it greatly accelerates the peptidogenomics method and thus facilitates a crucial step in the drug discovery pipeline. It consists of two main programs, each of which functions on a major class of peptidic natural products: Nrp2Path on non-ribosomal peptides (NRPs) and RiPP2Path on Ribosomally-synthesized and Post-translationally-modified Peptides (RiPPs).The Nrp2Path program probabilistically matches mass shift sequences or amino acid sequences of peptides to NRPS biosynthetic gene clusters. Pep2Path comes with a pre-configured database for Nrp2Path that contains data on all NRPS biosynthetic gene clusters in the most recent version of the GenBank database. Moreover, you can easily make your own databases from local files using the makedb program, which runs antiSMASH2 to identify NRPS gene clusters and store the predicted substrate specificities of their adenylation domains.
The RiPP2Path program matches mass shift sequences or amino acid sequences to the six-frame translation of genome sequences supplied in FASTA, GenBank or EMBL format.
Multiple nucleotide sequence files (or files that contain multiple nucleotide sequences themselves) can be analyzed at the same time.
The full version of Pep2Path, which is compatible with Linux, Windows and Mac, is available from the download area.
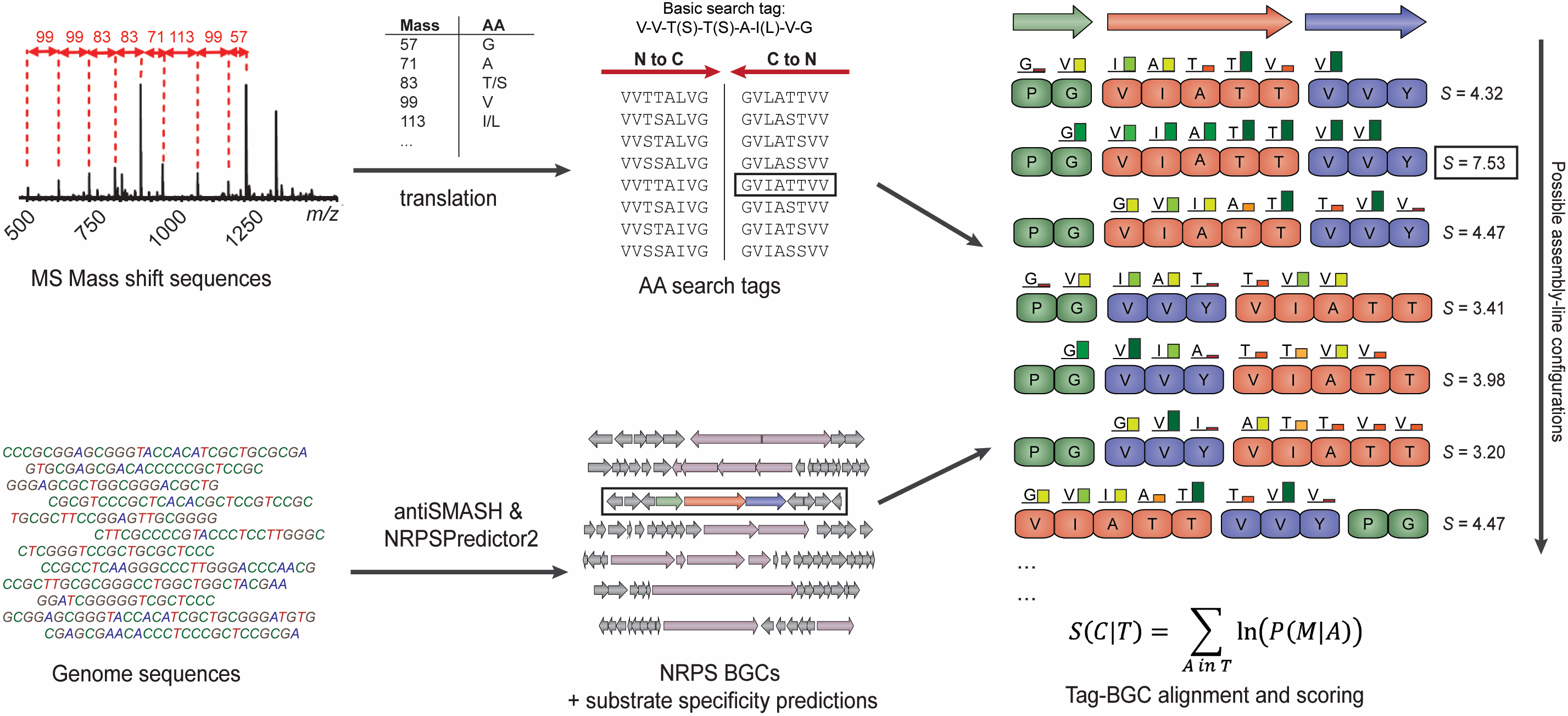
Overview of the Pep2Path process.